Welcome to OLIGAMI v2.00
OLIGAMI is a database of protein quaternary structures that can help us understand and interpret oligomer architecture at molecular interfaces.
OLIGAMI utilizes a formula based on an amino acid sequence (chain formula) to estimate the composition of oligomer chains in any biological complex
and allows you quick access to the coordinate data you want through the SCOPe
or CATH hierarchy. OLIGAMI also provides information concerning soluble and membrane proteins separately
using information from
the Orientations of Proteins in Membranes (OPM) database.
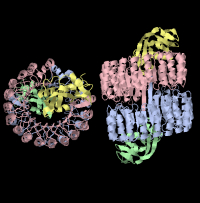
The Dimer interfaces are then classified based on their secondary structure features, providing a basis for browsing the oligomers. In the JSmol view of OLIGAMI, each oligomer subunit can be easily distinguished based on a color code.
Coordinate Files of the Biological Assembly of OLIGAMI
- Individual chain names So that each chain in JSmol view can be distinguished, each chain name is rewritten based on its individual name.
- Secondary structure definition All secondary structures in OLIGAMI coordinate files are recalculated with the program DSSP.
Chain Formula of OLIGAMI
- OLIGAMI utilizes three types of chain formulas The chain formulas express the chain composition of any biological assembly. OLIGAMI has three types of chain formulas for every entry, which distinguish the chains based on their amino acid sequences, their family sections of the SCOPe hierarchy, and their homology sections of the CATH hierarchy.
- Examples of chain formulas [A3][a] : Homotrimer with a short peptide
- Download a list of all chain formulas in OLIGAMI OLIGAMI-CF-v2.0-2022.txt
[AB](AB) : Heterodimer with a double-stranded DNA
[ ] : Polypeptide --- [AB] : Protein, [ab] : Small peptide
( ) : Nucleic acid --- (AB) : DNA, (ab) : RNA
Dimer Interface classification of OLIGAMI
- Homodimers and heterodimers composed of single-domain subunits are grouped hierarchically according to the secondary structure features of the interface. Please see the "Interfaces" page for details.
External Databases
Reference