Number of Protein Chains:29
- Classes: All alpha proteins
- Folds: Cytochrome c
- Superfamilies: Cytochrome c
- Families: Cytochrome bc1 domain
- Protein Domains: Cytochrome bc1 domain
- Species: Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932]
PDB Entries
| PDB ID | OPM | Mutant | Chain Formula | Resolution | HETNAM | ||
|---|---|---|---|---|---|---|---|
| Amino Acid sequence | SCOPe Family Level | CATH Homology Level | |||||
| 1ezv |  |
[A3B3C3D3E3F3G3H3I3JK] | [A6B3C3D3E3F3G3H3I2] | [A6B3C3D3E3F3G3H3I2] | 2.3 | FES:FE2/S2 (INORGANIC) CLUSTER HEM:PROTOPORPHYRIN IX CONTAINING FE SMA:STIGMATELLIN A UQ6:5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL | |
| 1kb9 |  |
[ABCDEFGHIJK] | [A2B2CDEFGHI] | [A2B2CDEFGHI] | 2.3 | CDL:CARDIOLIPIN FES:FE2/S2 (INORGANIC) CLUSTER HEM:PROTOPORPHYRIN IX CONTAINING FE PCF:1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE PEF:DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE PIE:1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL SMA:STIGMATELLIN A UMQ:UNDECYL-MALTOSIDE UQ6:5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL | |
| 1kyo |  |
[A2B2C2D2E2F2G2H2I2J2K2L] | [A4B4C2D2E2F2G2H2I2J] | [A4B4C2D2E2F2G2H2I2J] | 2.97 | FES:FE2/S2 (INORGANIC) CLUSTER HEC:HEME C M3L:N-TRIMETHYLLYSINE SMA:STIGMATELLIN A | |
| 1p84 |  |
[A2B2C2D2E2F2G2H2I2] | [A4B2C2D2E2F2G2H2] | [A4B2C2D2E2F2G2H2] | 2.5 | 3PE:1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE 3PH:1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE CDL:CARDIOLIPIN DBT:5-HEPTYL-6-HYDROXY-1,3-BENZOTHIAZOLE-4,7-DIONE FES:FE2/S2 (INORGANIC) CLUSTER HEC:HEME C PC1:1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE UMQ:UNDECYL-MALTOSIDE UQ6:5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL | |
| 2ibz | 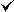 |
[ABCDEFGHIJK] | [A2B2CDEFGHI] | [A2B2CDEFGHI] | 2.3 | FES:FE2/S2 (INORGANIC) CLUSTER HEC:HEME C SMA:STIGMATELLIN A UQ6:5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL | |
| 3cx5 | 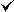 |
[A2B2C2D2E2F2G2H2I2J2K2L] | [A4B4C2D2E2F2G2H2I2J] | [A4B4C2D2E2F2G2H2I2J] | 1.9 | 6PH:(1R)-2-(PHOSPHONOOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYLPENTADECANOATE 7PH:(1R)-2-(DODECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYLTETRADECANOATE 8PE:(2R)-3-{[(S)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-2-(TETRADECANOYLOXY)PROPYL OCTADECANOATE 9PE:(1R)-2-{[(S)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(HEPTANOYLOXY)METHYL]ETHYL OCTADECANOATE CN3:(2R,5S,11R,14R)-5,8,11-TRIHYDROXY-2-(NONANOYLOXY)-5,11-DIOXIDO-16-OXO-14-[(PROPANOYLOXY)METHYL]-4,6,10,12,15-PENTAOXA-5,11-DIPHOSPHANONADEC-1-YL UNDECANOATE CN5:(5S,11R)-5,8,11-TRIHYDROXY-5,11-DIOXIDO-17-OXO-4,6,10,12,16-PENTAOXA-5,11-DIPHOSPHAOCTADEC-1-YLPENTADECANOATE FES:FE2/S2 (INORGANIC) CLUSTER FRU:BETA-D-FRUCTOFURANOSE GLC:ALPHA-D-GLUCOPYRANOSE HEM:PROTOPORPHYRIN IX CONTAINING FE M3L:N-TRIMETHYLLYSINE SMA:STIGMATELLIN A UMQ:UNDECYL-MALTOSIDE | |
| 3cxh | 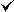 |
[A2B2C2D2E2F2G2H2I2J2K2L] | [A4B4C2D2E2F2G2H2I2J] | [A4B4C2D2E2F2G2H2I2J] | 2.5 | 6PH:(1R)-2-(PHOSPHONOOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYLPENTADECANOATE 7PH:(1R)-2-(DODECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYLTETRADECANOATE 8PE:(2R)-3-{[(S)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-2-(TETRADECANOYLOXY)PROPYL OCTADECANOATE 9PE:(1R)-2-{[(S)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(HEPTANOYLOXY)METHYL]ETHYL OCTADECANOATE CN6:(2R,5R,11S,14R)-2-(BUTANOYLOXY)-5,8,11-TRIHYDROXY-5,11-DIOXIDO-16-OXO-14-[(PROPANOYLOXY)METHYL]-4,6,10,12,15-PENTAOXA-5,11-DIPHOSPHANONADEC-1-YL UNDECANOATE FES:FE2/S2 (INORGANIC) CLUSTER FRU:BETA-D-FRUCTOFURANOSE GLC:ALPHA-D-GLUCOPYRANOSE HEM:PROTOPORPHYRIN IX CONTAINING FE M3L:N-TRIMETHYLLYSINE SMA:STIGMATELLIN A UMQ:UNDECYL-MALTOSIDE | |
*Chain Formula of OLIGAMI.
[ ] : Polypeptide - [AB] : Protein, [ab] : Small peptide
( ) : Nucleic acid - (AB) : DNA, (ab) : RNA
[ ] : Polypeptide - [AB] : Protein, [ab] : Small peptide
( ) : Nucleic acid - (AB) : DNA, (ab) : RNA
